Summary
A left ventricle (LV) simulation using a simplified ellipsoidal geometry. The cavity volume is equal to that of a Sprague-Dawley female rat. This simulates five cardiac cycles at which the LV produces a steady state pressure-volume (PV) loop. Instruction file found here.
Simulation Protocol
From the reference configuration, before the time loop begins, the left ventricular cavity volume is increased to approximately the end-diastolic value. This induces a passive stress response that yields a corresponding end-diastolic pressure. In this simulation, the initial diastolic loading is performed over 37 steps, with each step increasing the LV cavity volume by 0.004 mL.
After loading from the reference state, the time loop begins. During each time-step, the volume in each compartment of the Windkessel circulatory model is updated. Intracellular calcium concentration is then calculated via the three state paper calcium transient. This, along with deformation information is passed into MyoSim, where cross-bridge populations are calculated. Finally, a Newton solver calculates the new deformation to satisfy the balance of linear momentum. The simulation is run for 5 cycles, each at 170 ms, the cardiac period from data collected from rats.
Boundary Conditions & Assumptions
- LV base is fixed in the z-direction.
- Mean rigid body rotation and translation are zero.
- LV cavity volume is prescribed to match that solved for by a 3 compartment circulatory model.
- Contraction is driven by the three state kinetic scheme.
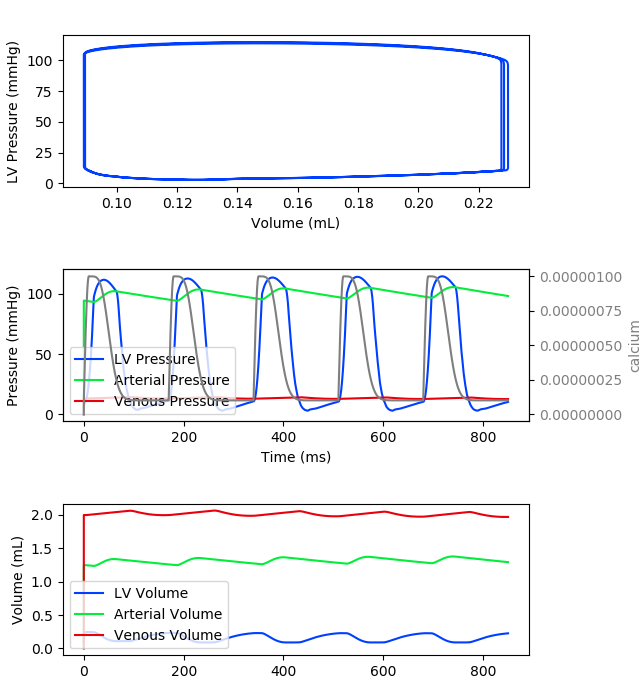
Results
The resulting pressure-volume loop, along with circulatory pressures, volumes and intracellular calcium are plotted below. These results can be plotted by executing
python /home/fenics/shared/source_code/plot_tools/viewpvloop.py _PV.txt
from the output directory.
The LV deformation can be seen below by applying the “Warp by Vector” filter to the “u_disp.pvd” files in the output directory using Paraview.